Advances in scalable DNA sequencing analysis: nf-core/sarek 3
- 06 May 2024
- Nina Gasparoni
Providing standardised, comparable, and reproducible omics workflows is a key goal for GHGA. Our bioinformatics team has been involved in the development and optimisation of several workflows for the research community.
In a new effort published in NAR Genomics and Bioinformatics - work led by co-Spokesperson Sven Nahnsen and colleagues - presents nf-core/sarek 3, a comprehensive variant calling and annotation pipeline designed for both germline and somatic samples.
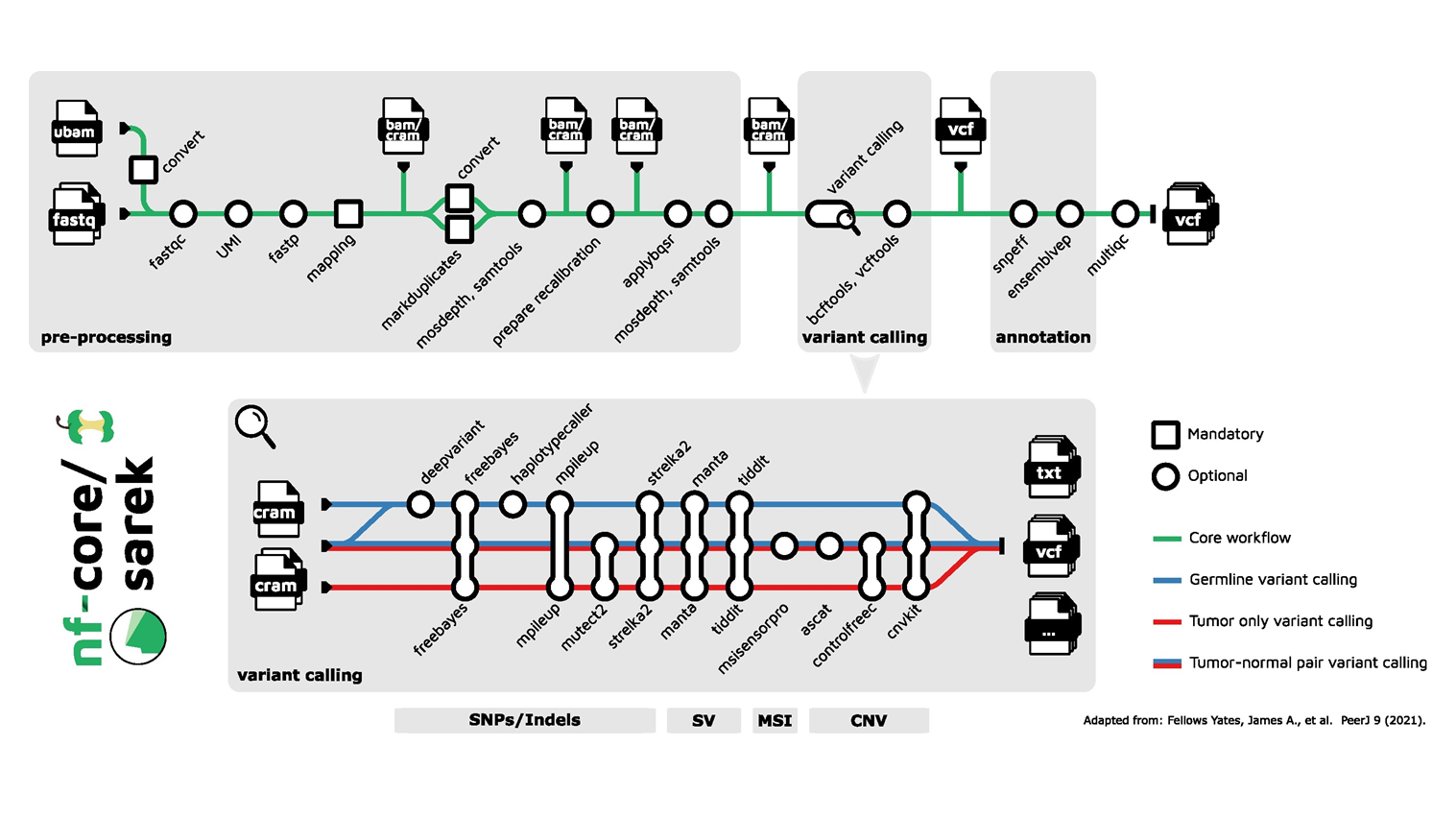
Understanding DNA variation is essential for several biomedical applications, particularly in cancer research and personalised medicine. The nf-core/sarek 3 pipeline addresses the growing need for highly scalable, portable, and automated workflows to process the vast amounts of sequencing data generated from thousands of samples. The original pipeline has undergone a major rewrite that has resulted in significant improvements to its performance. By leveraging the CRAM format and optimising intra-sample parallelization, the new version achieves significant reductions in storage requirements and compute costs.
The pipeline supports the analysis of single nucleotide variants (SNVs), small insertions and deletions (Indels), structural variations (SV) copy-number variations (CNVs) and micro-satellite instability (MSI). Its adaptability to different computing infrastructures, including commercial clouds and HPC clusters, ensures efficient large-scale and cross-platform data analysis while minimising costs and CO2 emissions.
GHGA's involvement in this project underscores our commitment to advancing accessible workflows that enable researchers worldwide to perform robust and cost-effective genomic analyses. By aligning with initiatives such as nf-core and refining existing workflows, GHGA continues to contribute to the advancement of genomic research and its applications in biomedicine.
Hanssen, F., Garcia, M. U., Folkersen, L., Pedersen, A. S., Lescai, F., Jodoin, S., Miller, E., Seybold, M., Wacker, O., Smith, N., Gabernet, G., & Nahnsen, S. (2024). Scalable and efficient DNA sequencing analysis on different compute infrastructures aiding variant discovery. NAR genomics and bioinformatics, 6(2), lqae031. https://doi.org/10.1093/nargab/lqae031