Benchmarking genomic variant calling
- 12 Oct 2023
- Nina Gasparoni, Christian Mertes
Accurate genomic variant calling is of utmost importance in the field of genomic research and diagnostics. To ensure precision and reliability in variant calling tools and workflows, continuous benchmarking against known standards is essential. This challenge has led to the development of a new approach that democratises benchmarking, making it accessible and transparent to all.
A recent publication, spearheaded by the Next Generation Sequencing Competence Network (NGS-CN) and GHGA, introduces NCBench, a platform for continuous benchmarking of genomic variant calling workflows.
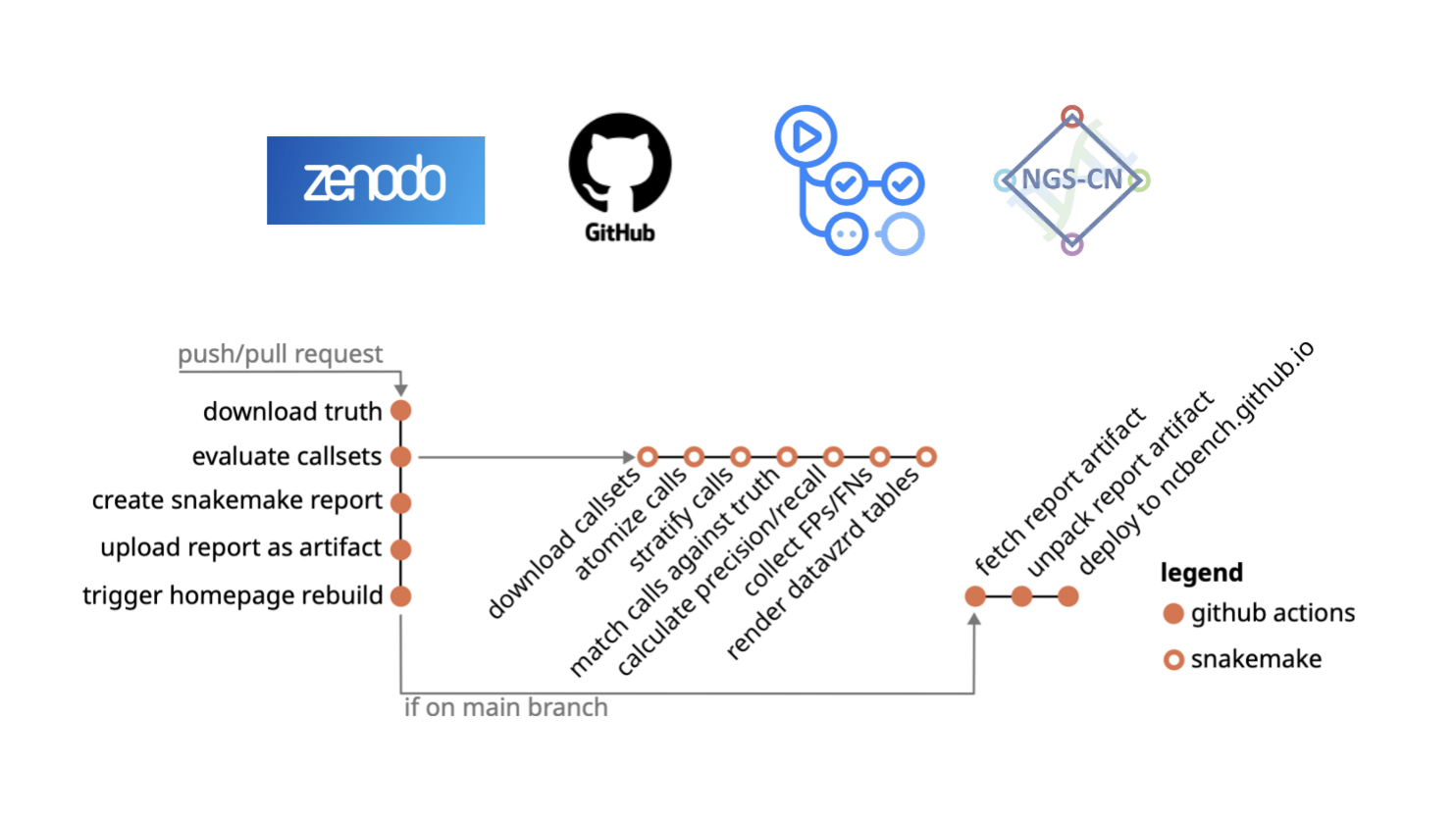
NCBench provides a comprehensive and reproducible benchmarking workflow for the evaluation of small genomic variant callsets in terms of recall, precision, and error patterns. Its continuous and open-source approach eliminates the need for specialised infrastructure, making benchmarking accessible and transparent to the research and diagnostics community but also to patients. By leveraging public platforms like GitHub, GitHub Actions, and Zenodo, NCBench streamlines benchmarking for researchers worldwide.
Evaluating over 40 callsets from various variant calling pipelines, NCBench uncovers subtle differences among callers, workflows, and datasets. This supports the developers in continuously extending and improving the workflows.
This open platform welcomes scientific contributions and fosters collaboration, reducing benchmark reimplementations and keeping tool performance up-to-date while harmonising and standardising benchmarking strategies.
Hanssen, F., Gabernet, G., Smith, N. H., Mertes, C., Neogi, A. G., Brandhoff, L., Ossowski, A., Altmueller, J., Becker, K., Petzold, A., Sturm, M., Stöcker, T., Sugirthan, S., Brand, F., Schmid, A., Buness, A., Probst, A.J., Motameny, S. & Köster, J. (2023). NCBench: providing an open, reproducible, transparent, adaptable, and continuous benchmark approach for DNA-sequencing-based variant calling. [version 1; peer review: awaiting peer review]. F1000Research, 12, 1125. https://doi.org/10.12688/f1000research.140344.1