The goal of the GHGA Metadata Model is to help our communities to richly describe their submitted genomic data and efficiently retrieve data of interest. In order to achieve this, we focus on making the metadata Findable, Accessible, Interoperable and Reusable (FAIR) by utilising established and widely used ontologies and vocabularies that we have defined in close collaboration with our partners.
The GHGA Metadata Model is implemented using the Linked Data Modelling Language LinkML and is openly accessible for everyone on the GHGA GitHub Repository. Here, one can track the changes made to the model, and access the schema and the autogenerated excel submission sheets.
The GHGA User Documentation provides detailed documentation about the GHGA Metadata Model and tools, including descriptions of the model itself, as well as the underlying concepts and standards.
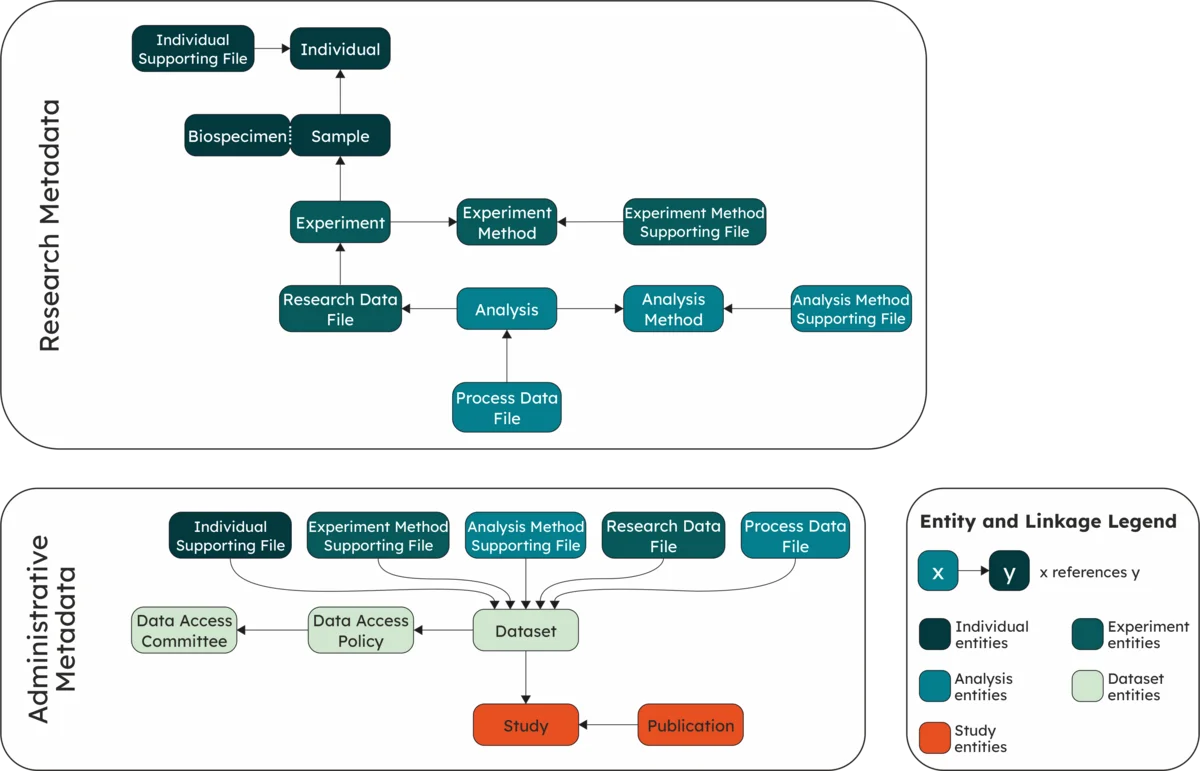
The model classes ensure interoperability and provide flexibility to capture different experiment and analysis types. The classes are grouped based on their functionality:
The Research Metadata focus on the reusability and FAIRness of the data and the Administrative Metadata capture the conditions and committee that govern data access.
Research Metadata
Administrative Metadata
The data can be submitted using GHGAs submission spreadsheet, which reflects the GHGA Metadata Model and enables the GHGA Data Portal to display valuable information about a submitted dataset through the linkage of all categories. Richly described metadata will help to promote data submitters datasets and encourage the community to reuse the data.