Major bioinformatic workflow releases with GHGA contribution
- 29 Jul 2022
- Nicholas Smith
Within the GHGA workflow workstream, our bioinformatics team has been making steady progress in our effort to provide standardized, comparable, and reproducible omics workflows for the research community. Recently, GHGA was involved in the releases of three workflows (sarek 3.0, nanoseq 3.0 und DROP 1.2) which - alongside ongoing efforts, we highlight here.
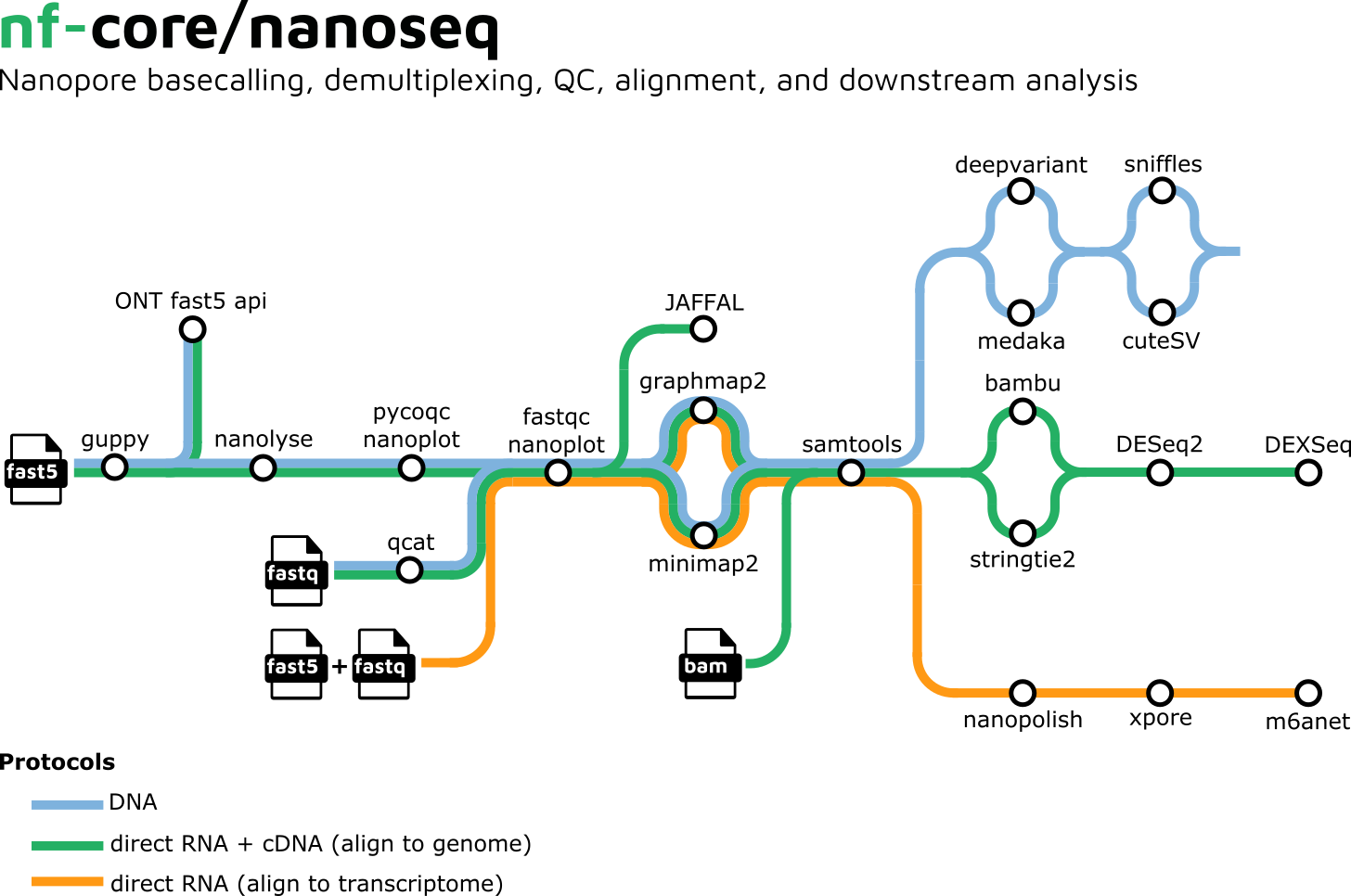
In conjunction with the nf-core developers, GHGA bioinformaticians have worked to push sarek 3.0 to be released! This new version is a completely modular workflow that is agnostic of a compute environment and includes dozens of community tools that offer germline and tumor variant calling from start to finish. The team has also worked together with the long read sequencing community to improve and extend the nanoseq workflow. The new major release nanoseq 3.0 includes now DNA variant calling alongside RNA modification and fusion detection. The new features are discussed in the bytesize video here.
Additionally, in conjunction with the Gagneur lab at TUM we have released a new version of the Detection of RNA Outlier Pipeline (DROP 1.2). DROP, which is already used by the European Solve-RD project, focuses on the rare transcriptomic events of aberrant splicing, expression, and the multi-omics approach of monoallelic expression. We reported about an earlier release and landmark paper in April here.
In addition to these milestones, the GHGA workflow workstream is currently partnering with the developers of nf-core scrnaseq 2.0 and scatac to create high-quality standardized and reproducible workflows incorporating single-cell RNA-seq and ATAC-seq datasets. GHGA is also involved in the benchmarking efforts of the German Next Generation Sequencing Competence Network (NGS-CN) looking to standardize the benchmarking process for sequencing centers across Germany.
We look forward to continuing to create usable products, and appreciate community engagement! Please fill out the survey to help guide the products and workflows GHGA develops and provides for users: https://www.ghga.de/news/detail/ghga-survey-for-community-driven-development
Open Source Access to all workflows can be found on Github:
https://github.com/nf-core/sarek
https://github.com/nf-core/nanoseq