Designing software is half science and half art. At GHGA, this is not different, since we have to find a creative balance between many aspects and requirements:
On one hand, we would like to have a first version of our genome archive operational very soon. On the other hand, GHGA is a project that shall run for 10 years and beyond, so long-term maintainability and extensibility is a key consideration. Our software solutions should be straight forward to deploy to simplify the onboarding of new data centers to our network. At the same time, we have to provide the needed flexibility to adapt to the resources and infrastructures that are locally available. Moreover, the tools that we are developing should not only be used by us, but we would also like to serve the broader national and international research and health care community.
Everything starts with the right development culture. Continuously optimizing our agile development processes goes thereby hand in hand with DevOps practices which make us think of software development and operation as one unit. This is why we closely coordinate with the Data Hub Operations Workstream. Moreover, choosing progressive yet robust architecture patterns is another key aspect of tackling the above challenges. This is why, from day one, we are implementing a domain-driven microservice architecture. This is not only easy to maintain, but it also facilitates major refactorings that will be necessary when the scope of our projects changes over time. Furthermore, to be independent of a specific cloud provider and to enable frictionless continuous deployment, we rely on the container orchestration-solution Kubernetes and its associated ecosystem. Finally, we put a lot of effort into aligning with national and international software standards and we aim to actively push their development forward by participating in community efforts of the NFDI, ELIXIR Europe and the GA4GH.
Achievements & Products
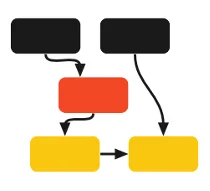
The federated nature of GHGA requires a localized expertise when it comes to the production level operations. There are two main themes of these operations, (1) Data Stewardship and (2) DevOps operations. GHGA production work will require close collaboration of these two groups for running day to day GHGA activities.
Data Stewardship. One topic the Operations Workstream focuses on is the details of the helpdesk structure and mode of operation. Data stewards at each hub will form the GHGA helpdesk and support users with data submission and data access requests. Close collaboration with the sequencing centers ensures tight connections to GHGA’s major data submitters.
DevOps. In the first phase of GHGA, DevOps operations are carried out together with the Software & Infrastructure Workstream (see below). This way, the deployment and operations strategy will be closely aligned with software development.
All processes within the Data Hub Operations Team are organized using standard operating procedures (SOPs), which are a vital tool for making sure that data hubs work in a reproducible and safe manner individually and jointly.
The ELSI (Ethical, Legal, and Social Implications) team consists of legal scholars and ethics researchers who are working in close collaboration to develop the foundations for the ethical and legal context of GHGA. Together we provide the necessary ethical and legal documents for GHGA (e.g., consent forms, governance papers, and policy documents), aim to ensure GHGA’s legal implementation and interoperability and explore strategies to involve patients and data subjects in the conception and governance of GHGA in order to establish broad and lasting societal support for the project.
The ethics team has been working on a consent tool to allow data providers to share their data with GHGA in the future and on a guide on how consent modules should be used to extend already existing consent forms for GHGA’s purposes. Moreover, we are working with patient representatives to develop informational resources for patients and gather input on ethical and regulatory issues. The result will be a white paper describing how patients can be involved in GHGA governance and thus help to build and keep trust with stakeholders.
The legal team has been focusing on the legal basis for data processing and legacy consent (i.e., consent obtained in the past). We are also working on risk assessment, de-identification and anonymization methods and a potential Code of Conduct to implement the governance framework for GHGA and enhance its legal interoperability for data processing within the EU and international data spaces.
Achievements & Products
GHGA is embedded in an ecosystem of national and international efforts. Ensuring that GHGAs solutions for the secure sharing of human omics data align with initiatives such as GDI, fEGA and genomDE is a goal across our workstreams.
Our GDI workstream is dedicated to the alignment with the European Genomic Data Infrastructure (GDI) - a key project within GHGAs alignment efforts. The GDI initiative aims to create a cross-border interconnected network of national genome collections to fulfil the goal of the 1+ Million Genomes (1+MG) initiative. Serving as the german node within GDI, our dedicated GDI workstream ensures that the infrastructure developed with GHGA is compatible with GDI nodes across Europe.
Within the GDI workstream we focus on aligning both technical and metadata aspects. On the technical side, we're integrating the GDI Starter Kit, which offers essential services such as data discovery, access management, secure storage, and processing, with our existing GHGA services. This means linking our data access management and secure storage with GDI's data discovery and processing services. We are currently exploring the use of Beacon v2 for data discovery within GHGA. On the metadata side, we're ensuring alignment with GDI's centralised user portal, which aggregates metadata from the various nodes for dataset search.
Achievements & Products
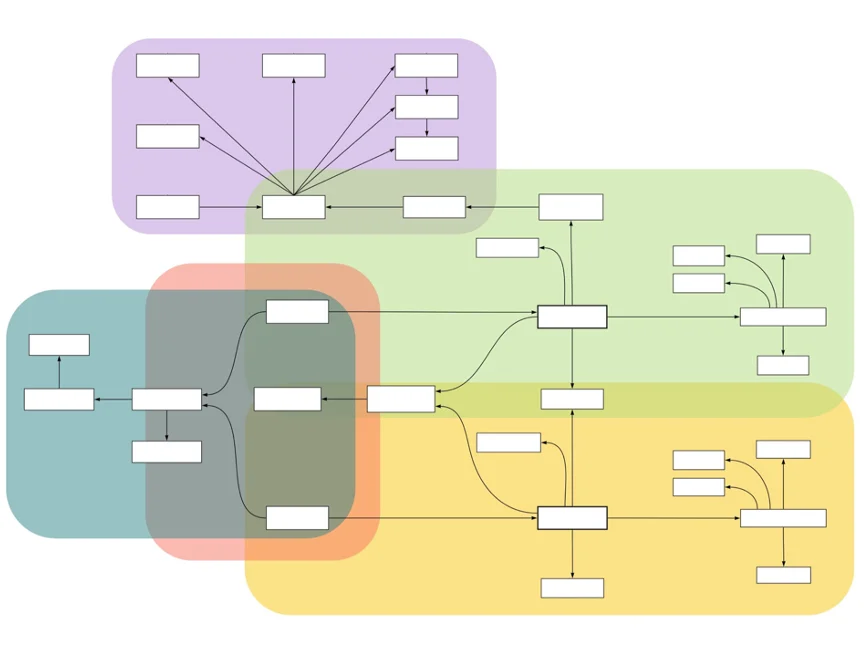
The Metadata Workstream provides the model for the data stored in GHGA. It is a joint effort of the conceptual and technical departments of GHGA. The team brings together experts with extensive knowledge from various areas that feed into the definition of the metadata schema, which serves as the framework for the GHGA Metadata Model.
The starting point of the workstream was the evaluation of already existing and well-established metadata models, with a focus on the European Genome-phenome Archive (EGA) as well as the cancer and rare disease communities. With the prior knowledge from various portals, a prototype schema was set up and refined over several rounds of feedback and testing. The GHGA Metadata Schema is openly accessible on GitHub
We make GHGA’s metadata FAIR by utilizing established and widely used ontologies and vocabularies that help our communities to describe their submitted data as well as to retrieve data of interest. All ontologies and vocabularies are evaluated based on their maintenance and richness of content with the help of https://fairsharing.org. The identified metadata, ontologies and vocabularies were structured in our metadata schema, which is technically implemented using the Linked data Modelling Language. LinkML helps us to create and update the metadata schema in one place and provides GHGA’s technical stack with definitions of the schema in different modeling languages, such as JSON and RDF.
Achievements & Products

At GHGA, data scientists, biomedical researchers and clinicians from more than 20 institutions are working together to bring this ambitious project to life. This results in a very interdisciplinary workforce of over 80 members who are involved in the diverse working areas of GHGA.
To ensure that GHGA is running smoothly, the Project Management Team strives to universally support the overall team and the workstreams. This includes administrative tasks like finances and recruiting. Additionally, the Project Management Team is supporting the workstreams in their general organisation (including reporting), the development of the legal framework, and organisation of internal and external meetings. Furthermore, the Project Management Team is involved in the governance of the project, organising regular meetings with the Board of Directors and the Scientific Advisory Board.
Being at the interface between the GHGA consortium and the NFDI, the Project Management Team also participates at various levels in the different NFDI working groups.
Achievements & Products

GHGA’s communication channels are diverse. Reaching different audiences with the same message: Data sharing in omics research is safe, if all necessary safety precautions are taken (which we do!), and important to drive scientific discovery.
Building the infrastructure with research and clinical communities in mind, GHGA is in close contact with the omics data producers and users. Only when we know what the communities need can we deliver a platform that serves all of their requirements.
Presenting at conferences and holding workshops, we aim to not simply advertise GHGA but also to encourage the principles of FAIR data sharing. Data sharing is a collaborative effort. Between scientists and clinicians. But also between different initiatives, making sure national and international efforts are aligned and ideally follow similar standards. GHGA aims to connect genome research undertaken in German institutions.
GHGA is passionate about engaging with the public to increase understanding and awareness around omic research and ultimately enabling informed decisions on data sharing. Genome research can be life saving, is interesting and all around us. Making this visible for everyone, we are seeking a dialogue with the public by exploring different approaches, such as local events and our podcast ‘Der Code des Lebens’.
Achievements & Products
In the training team we are passionate about supporting our users and communities in all topics relevant to sharing data in the field of biomedical research and health care, as well as related bioinformatic methods. Therefore we made it our mission to provide training and learning opportunities in these areas.
Our material ranges from ELSI topics on consent and consent tools, to technical topics around metadata, bioinformatic analysis of DNA and RNA to Good Practice in study design and statistical analysis. Together with the GHGA outreach team, we also aim to inform our users - and an interested wider audience - on topics close to our hearts, e.g. FAIR data sharing and general research data management, by providing courses and additional material on these interesting topics.
An important part of GHGA’s mission is to facilitate sharing of genomic (and other omics) data from different data providers. To make this possible, data needs to be uploaded into the GHGA Data Portal. Once safely stored there, data can be browsed, downloaded (after successful application for permission) and, with increasing functionality of GHGA services, even analysed using dedicated workflows developed by GHGA and the community.
The process of uploading data into such a service can be daunting as it includes pre-submission data preparation, upload of accompanying metadata and knowledge of how to technically upload the data. Also, when browsing uploaded data, finding the right data sets for your analysis and subsequently applying for permission to download these can be aided by providing guidance by the creators of the platform.
Making these user experiences as smooth as possible is one of the major goals of the GHGA Training workstream. Therefore, we will provide training material on how to navigate all aspects of the data portal. At later stages of development - when workflows for data analysis and other resources have been added - training and support material for these will also be provided. Training will come in different flavours: you can choose from live webinars, on-demand video tutorials or text-based materials - whichever suits you best.
Achievements & Products
Course FAIR in (biological) practice: https://www.youtube.com/playlist?list=PLXMwmQxyLByV8wQ4i9NwG_cDaJGc5Nrhj
Webinare:
Standardized NGS Workflows at GHGA
At GHGA, the Workflow Workstream is dedicated to standardizing and harmonizing Next Generation Sequencing (NGS) analysis workflows to support high-quality, FAIR-compliant data sharing across the German life science research community. Rather than building new workflows from scratch, we enhance and extend existing community-driven solutions, closely aligning with established standards such as GA4GH, nf-core, and Snakemake.
Our goal is to enable the uniform processing of raw sequencing data into ready-to-use research outputs, ensuring consistency and comparability across all submitted datasets.
Enabling Reliable Cross-Study Analysis
Standardized workflows are essential for reliable cross-study analyses. To achieve this, GHGA plans to implement automated quality control (QC) pipelines for all data submitted to the GHGA archive. Once in place, these pipelines will generate data quality reports accessible through the GHGA data portal. These reports will enable users to filter and query datasets based on quality criteria tailored to their specific research needs.
Ensuring Quality and Reproducibility
Data quality and reproducibility are at the core of our workflow strategy. Each pipeline is subjected to rigorous continuous integration and deployment (CI/CD) testing to ensure technical stability. We also benchmark biological performance using both synthetic and experimental datasets, such as Seracare synthetic samples, CHM cell lines, and Genome in a Bottle (GiaB) references.
As we develop deployable and scalable pipelines, we optimize resource usage—tuning parameters based on provenance and trace data to minimize computational overhead, particularly for large-scale data processing.
Supporting a FAIR Data Ecosystem
Our efforts extend beyond pipeline development. We are actively contributing to a more robust FAIR data ecosystem by:
All GHGA workflows are open-source and registered on community platforms, making them Findable, Accessible, Interoperable, and Reusable—key principles guiding GHGA’s transformation from a secure data archive into a fully integrated, national data service provider.
More information on our community-driven projects and analysis pipelines can be found here.